Pathologists agreed just three-quarters of the time when diagnosing breast cancer from biopsy specimens, according to a recent study.
The difficult, time-consuming process of analyzing tissue slides is why pathology is one of the most expensive departments in any hospital.
Faisal Mahmood, assistant professor of pathology at Harvard Medical School and the Brigham and Women’s Hospital, leads a team developing deep learning tools that combine a variety of sources — digital whole slide histopathology data, molecular information, and genomics — to aid pathologists and improve the accuracy of cancer diagnosis.
Mahmood, who heads his eponymous Mahmood Lab in the Division of Computational Pathology at Brigham and Women’s Hospital, spoke this week about this research at GTC DC, the Washington edition of our GPU Technology Conference.
The variability in pathologists’ diagnosis “can have dire consequences, because an uncertain determination can lead to more biopsies and unnecessary interventional procedures,” he said in a recent interview. “Deep learning has the potential to assist with diagnosis and therapeutic response prediction, reducing subjective bias.”
Depending on the type of cancer and the pathologist’s level of experience, it can take 15 minutes or more for a pathologist to analyze a biopsy slide. If a single patient has a couple dozen slides, it can add up quick.
And to decide on a treatment plan, doctors also take into account other data sources like patient and familial medical history, as well as molecular and genomic data when it’s available.
Mahmood’s team uses NVIDIA GPUs on premises and in the cloud to develop its AI tools for pathology image analysis that incorporates all of these data sources.
“By working with whole slide images and fusing multimodal data sources we are algorithmically moving closer and closer to the clinical workflow,” Mahmood said. “This will enable us to run prospective studies with AI-assisted pathology diagnosis tools that use multimodal data.”
AI Sees the Big Picture
Digitized whole slide images taken during a tissue biopsy are huge — each can be more than 100,000 by 100,000 pixels. To efficiently compute with such large files, deep learning developers often choose to chop a slide into individual patches, making it easier for a neural network to process. But this tactic makes it incredibly time-consuming for researchers to hand-label the training data.
The Mahmood Lab is developing deep learning models that parse whole tissue slides at once in a data-efficient method, using NVIDIA GPUs to accelerate training and inference of their neural networks. These models can be used for patient selection and stratification into treatment groups for precision therapies.
For prototyping their deep learning models, and for inference, the team relies on four on-prem machines with NVIDIA GPU clusters. To train graph convolutional networks and contrastive predictive coding models with large pathology images, the researchers use NVIDIA V100 Tensor Core GPUs in Google Cloud.
“The modern GPU is what gives us the ability to train deep learning models on whole slides,” said Max Lu, a researcher in the Mahmood Lab. “The benefit is that it doesn’t require modifying the current clinical workflow, because pathologists are analyzing and preparing reports for whole slides anyways.”
Joining Sources
Pathologists often make their determinations using a wealth of data ranging from tissue slides, immunohistochemistry markers and genomic profiles. But most current deep-learning based diagnosis methods rely on a single data source or on trivial methods of fusing information.
This led Mahmood Lab researchers to develop mechanisms that combine microscope and genomic data in a much more heuristic and holistic manner. Initial results suggest that adding information from genomic profiles and graph convolutional networks can improve diagnostic and prognostic models.
Sliding into the Pathology Workflow
Mahmood sees two potential ways in which deep learning could be incorporated into pathologists’ workflow. AI-annotated slide images could be used as a second opinion for pathologists to help improve the quality and consistency of diagnoses.
Or, computational pathology tools could screen out all the negative cases, so that pathologists only need to review biopsy slides that are likely positive, significantly reducing their workloads. There’s a precedent for this: In the 1990s, hospitals began using third-party companies to scan and stratify pap smear slides, throwing out all the negative cases.
“If there are 40,000 breast cancer tissue slides and 20,000 are negative, that half would be stratified out and the pathologist wouldn’t see it,” Mahmood said. “Just by reducing the pathologist’s burden, variability is likely to go down.”
To test and validate their algorithms, the researchers plan to conduct retrospective and prospective studies using biopsy data from the Dana Farber Cancer Institute. They will study whether a pathologist’s analysis of a biopsy slide changes after seeing the algorithm’s determination — and whether using AI reduces variation in diagnosis.
Mahmood Lab researchers will present their deep learning projects at the NeurIPS conference’s ML4H workshop in December.
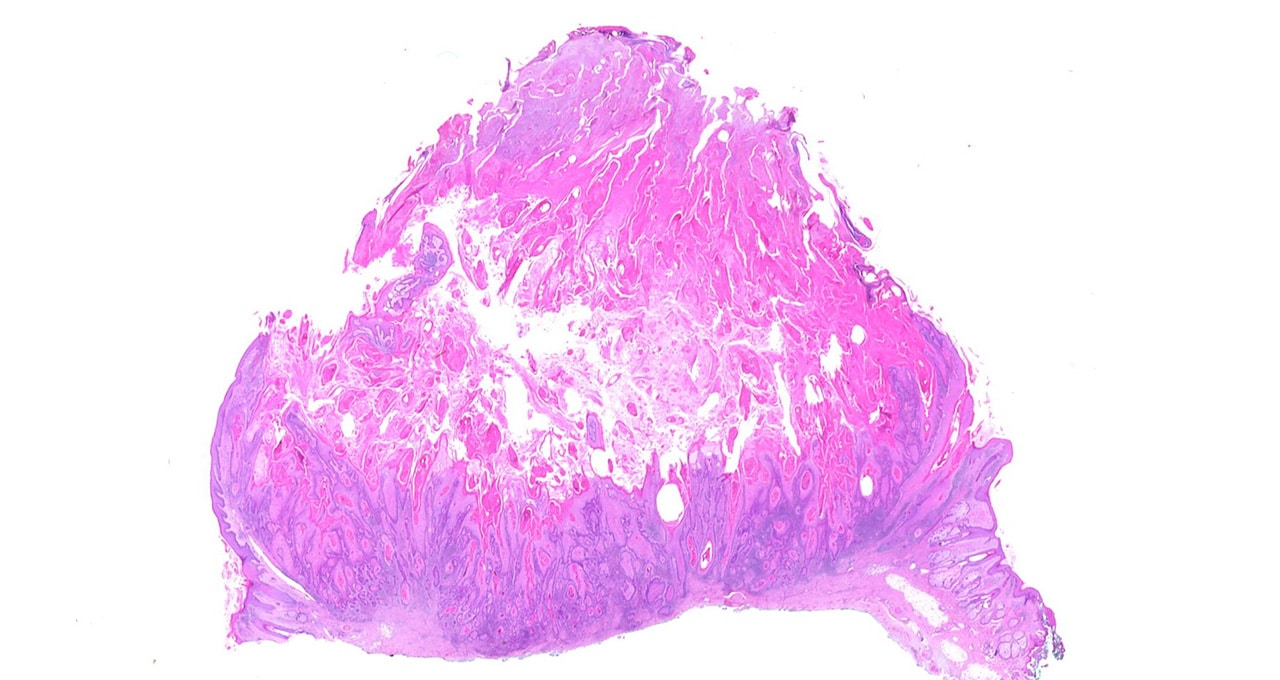
Main image shows a whole slide of keratocanthoma, a type of skin tumor. Image by Alex Brollo, licensed from Wikimedia Commons under CC BY-SA 3.0.