Days after giving an online talk on how GPUs reduce the time to understand diseases, Margaret Linan felt sick, and awoke at 3am gasping for air.
“It was quite frightening,” said Linan, a computational research scientist at Mount Sinai’s Icahn School of Medicine, in New York. With help from first responders, she got her breath back in about 20 minutes.
Her symptoms diminished over a few days, but for a time, it was unsettlingly strange.
“I’m pretty sure it was a minor version of the coronavirus,” said Linan, who had been sheltering at home.
It was a poignant reminder of the value of her work. Linan develops and tests software that accelerates the discovery of mutations in human genomes, used to identify treatments for diseases such as cancer.
At GTC Digital, she presented her latest results, showing dramatic speedups in genomics using a mix of GPUs and CPUs. Her analysis showed how NVIDIA’s RAPIDS software for accelerating data science on GPUs eased the transition from today’s mainly CPU-based programs.
6x Speedups and More in Genomics
Processing and analyzing genome samples can take months on a CPU. Doctors with cancer patients sometimes depend on such studies for life-saving treatments.
In Linan’s experiments, GPUs accelerated one genomic analysis by more than 6x. Another job delivered even greater savings due to the high number of number of runs it required.
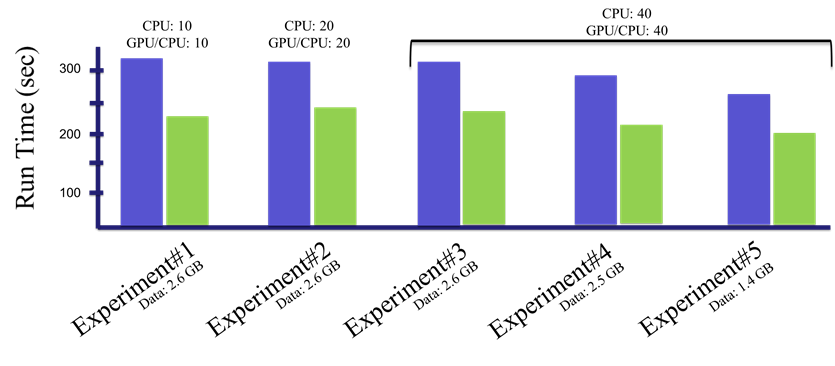
A server with a single GPU in her lab handled initial tests. More mature experiments ran on Mount Sinai’s Minerva system that packs 48 NVIDIA V100 Tensor Core GPUs, up to four available for a single job. Advanced tests that used eight V100 GPUs were sent to the AWS cloud.
Comparisons of six GPU software frameworks favored RAPIDS. Bioinformatics specialists familiar with Python programs it supports find it easiest to use.
“You feed in an object, and it does what you expect it will,” she said.
Open Source GPU Code on the Way
Mount Sinai’s researchers increasingly tap into GPUs on the Minerva system or in the cloud to handle their toughest jobs, Linan reported.
That’s one reason why she aims to create a suite of open source programs to accelerate genomics in GPUs. She’s almost done creating a GPU-accelerated version of the Genome Analysis Toolkit, one of about five key tools in the field, and has made progress on two others.
“By the end of the year, I hope to have beta versions available for our investigators to test,” Linan said.
Use cases for AI are growing among researchers at the New York hospital.
Late next year, Mount Sinai plans to open a $100 million research center for AI in healthcare. It will extend the mission of its existing Institute for Genomic Health that leverages AI.
Deep Patient, another project at the medical school, uses neural networks to predict which patients are at most risk of developing diabetes or certain cancers. In addition, Mount Sinai has spawned two startups in healthcare AI. RenalytixAI designs clinical diagnostics for kidney disease; Sema4 uses AI to develop personalized cancer care programs.
“GPUs will be leveraged on many large-scale research projects for many years to come,” she said.
