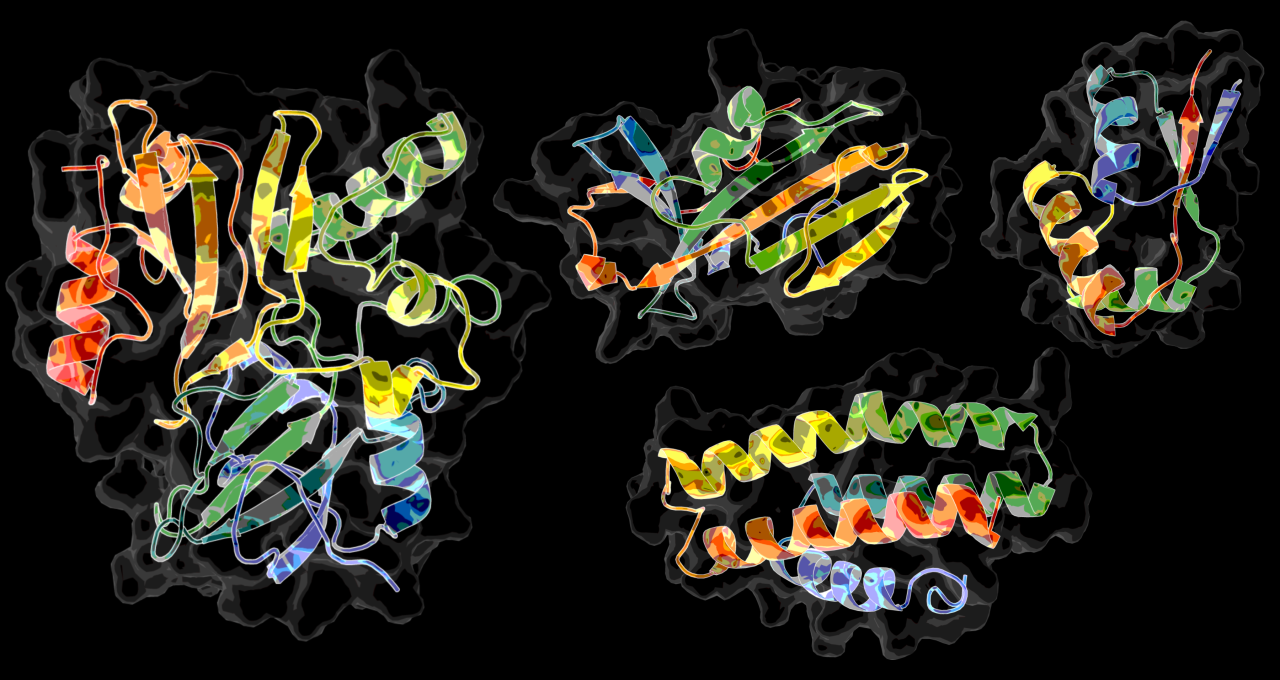
Using AI and a supercomputer simulation, Ken Dill’s team drew the equivalent of wanted posters for a gang of proteins that make up COVID-19. With a little luck, one of their portraits could identify a way to arrest the coronavirus with a drug.
When the pandemic hit, “it was terrible for the world, and a big research challenge for us,” said Dill, who leads the Laufer Center for Physical & Quantitative Biology at Stony Brook University, in Long Island, New York.
For a decade, he helped the center assemble the researchers and tools needed to study the inner workings of proteins — complex molecules that are fundamental to cellular life. The center has a history of applying its knowledge to viral proteins, helping others identify drugs to disable them.
“So, when the pandemic came, our folks wanted to spring into action,” he said.
AI, Simulations Meet at the Summit
The team aimed to use a combination of physics and AI tools to predict the 3D structure of more than a dozen coronavirus proteins based on lists of the amino acid strings that define them. It won a grant for time on the IBM-built Summit supercomputer at Oak Ridge National Laboratory to crunch its complex calculations.
“We ran 30 very extensive simulations in parallel, one on each of 30 GPUs, and we ran them continuously for at least four days,” explained Emiliano Brini, a junior fellow at the Laufer Center. “Summit is a great machine because it has so many GPUs, so we can run many simulations in parallel,” he said.
“Our physics-based modeling eats a lot of compute cycles. We use GPUs almost exclusively for their speed,” said Dill.
Sharing Results to Help Accelerate Research
Thanks to the acceleration, the predictions are already in. The Laufer team quickly shared them with about a hundred researchers working on a dozen separate projects that conduct painstakingly slow experiments to determine the actual structure of the proteins.
“They indicated some experiments could be done faster if they had hunches from our work of what those 3D structures might be,” said Dill.
Now it’s a waiting game. If one of the predictions gives researchers a leg up in finding a weakness that drug makers can exploit, it would be a huge win. It could take science one step closer to putting a general antiviral drug on the shelf of your local pharmacy.
Melding Machine Learning and Physics
Dill’s team uses a molecular dynamics program called MELD. It blends physical simulations with insights from machine learning based on statistical models.
AI provides MELD key information to predict a protein’s 3D structure from its sequence of amino acids. It quickly finds patterns across a database of atomic-level information on 200,000 proteins gathered over the last 50 years.
MELD uses this information in compute-intensive physics simulations to determine the protein’s detailed structure. Further simulations then can predict, for example, what drug molecules will bind tightly to a specific viral protein.
“So, both these worlds — AI inference and physics simulations — are playing big roles in helping drug discovery,” said Dill. “We get the benefits of both methods, and that combination is where I think the future is.”
MELD runs on CUDA, NVIDIA’s accelerated computing platform for GPUs. “It would take prohibitively long to run its simulations on CPUs, so the majority of biological simulations are done on GPUs,” said Brini.
Playing a Waiting Game
The COVID-19 challenge gave Laufer researchers with a passion for chemistry a driving focus. Now they await feedback on their work on Summit.
“Once we get the results, we’ll publish what we learn from the mistakes. Many times, researchers have to go back to the drawing board,” he said.
And every once in a while, they celebrate, too.
Dill hosted a small, socially distanced gathering for a half-dozen colleagues in his backyard after the Summit work was complete. If those results turn up a win, there will be a much bigger celebration extending far beyond the Stony Brook campus.