For researchers like Max Zimmerman, it was a welcome pile-on to tackle a global pandemic.
A million citizen scientists donated time on their home systems so the Folding@home consortium could calculate the intricate movements of proteins inside the coronavirus. Then a team of NVIDIA simulation experts combined the best tools from multiple industries to let the researchers see their data in a whole new way.
“I’ve been repeatedly amazed with the unprecedented scale of scientific collaborations,” said Zimmerman, a postdoc fellow at the Washington University School of Medicine in St. Louis, which hosts one of eight labs that keep the Folding@home research network humming.
As a result, Zimmerman and colleagues published a paper on BioRxiv, showing images of 17 weak spots in coronavirus proteins that antiviral drug makers can attack. And the high-res simulation of the work continues to educate researchers and the public alike about the bad actor behind the pandemic.
“We are in a position to make serious headway towards understanding the molecular foundations of health and disease,” he added.
An Antiviral Effort Goes Viral
In mid-March, the Folding@home team put many long-running projects on hold to focus on studying key proteins behind COVID. They issued a call for help, and by the end of the month the network swelled to become the world’s first exascale supercomputer, fueled in part by more than 280,000 NVIDIA GPUs.
Researchers harnessed that power to search for vulnerable moments in the rapid and intricate dance of the folding proteins, split-second openings drug makers could exploit. Within three months, computers found many promising motions that traditional experiments could not see.
“We’ve simulated nearly the entire proteome of the virus and discovered more than 50 new and novel targets to aid in the design of antivirals. We have also been simulating drug candidates in known targets, screening over 50,000 compounds to identify 300 drug candidates,” Zimmerman said.
The coronavirus uses cunning techniques to avoid human immune responses, like the Spike protein keeping its head down in a closed position. With the power of an exaflop at their disposal, researchers simulated the proteins folding for a full tenth of a second, orders of magnitude longer than prior work.
Though the time sampled was relatively short, the dataset to enable it was vast.
The SARS-CoV-2 spike protein alone consists of 442,881 atoms in constant motion. In just 1.2 microseconds, it generates about 300 billion timestamps, freeze frames that researchers must track.
Combined with the two dozen other coronavirus proteins they studied, Folding@home amassed the largest collection of molecular simulations in history.
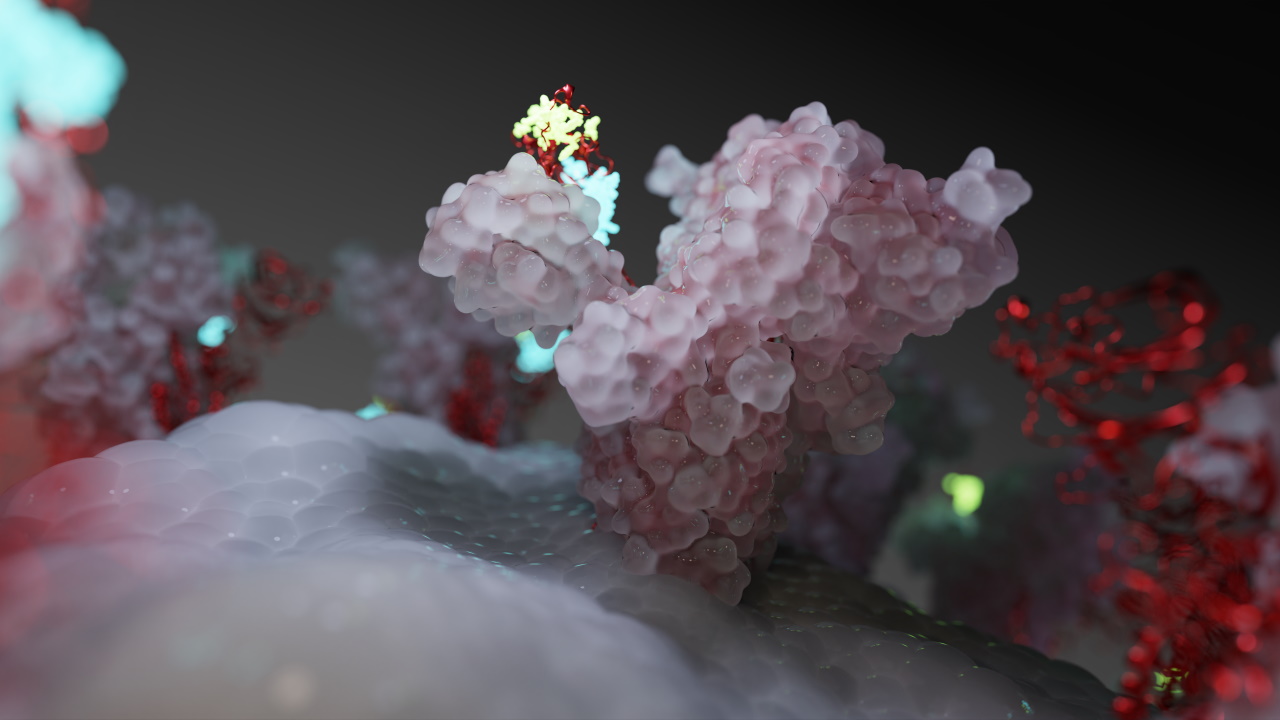
Omniverse Simulates a Coronavirus Close Up
The dataset “ended up on my desk when someone asked what we could do with it using more than the typical scientific tools to really make it shine,” said Peter Messmer, who leads a scientific visualization team at NVIDIA.
Using Visual Molecular Dynamics, a standard tool for scientists, he pulled the data into NVIDIA Omniverse, a platform built for collaborative 3D graphics and simulation soon to be in open beta. Then the magic happened.
The team connected Autodesk’s Maya animation software to Omniverse to visualize a camera path, creating a fly-through of the proteins’ geometric intricacies. The platform’s core technologies such as NVIDIA Material Definition Language (MDL) let the team give tangible surface properties to molecules, creating translucent or glowing regions to help viewers see critical features more clearly.
With Omniverse, “researchers are not confined to scientific visualization tools, they can use the same tools the best artists and movie makers use to deliver a cinematic rendering — we’re bringing these two worlds together,” Messmer said.
Simulation Experts Share Their Story Live
The result was a visually stunning presentation where each spike on a coronavirus protein is represented with more than 1.8 million triangles, rendered by a bank of NVIDIA RTX GPUs.
Zimmerman and Messmer will co-host a live Q&A technical session Oct. 8 at 11 AM PDT to discuss how they developed the simulation that packs nearly 150 million triangles to represent a millisecond in a protein’s life.
The work validates the mission behind Omniverse to create a universal virtual environment that spans industries and disciplines. We’re especially proud to see the platform serve science in the fight against the pandemic.
The experience made Zimmerman “incredibly optimistic about the future of science. NVIDIA GPUs have been instrumental in generating our datasets, and now those GPUs running Omniverse are helping us see our work in a new and vivid way,” he said.
Visit NVIDIA’s COVID-19 Research Hub to learn more about how AI and GPU-accelerated technology continues to fight the pandemic. And watch NVIDIA CEO Jensen Huang describe in a portion of his GTC keynote below how Omniverse is playing a role.